*Corresponding Author:
Ana Paula Ferreira Costa,
Universidade Federal do Rio Grande do Norte, Departamento de Análises Clínicas e Toxicológicas, Natal, RN, Brazil
Tel: +55 8433429829
Fax: +55 8433429796
E-mail: ana-paula-rf@hotmail.com
Abstract
Human leukocyte antigen (HLA)-G molecule acts as a potential factor in the regulation of immune responses and its expression in virus-infected cells may enable the virus to escape immunosurveillance. The HLA-G 14-base pairs (14-bp) insertion/deletion (ins/del) polymorphism has been associated with the magnitude of HLA-G production. Here, we have studied the association of 14-bp ins/del polymorphism with the risk of cytomegalovirus (CMV) infection in renal transplant patients. One hundred and six kidney-transplanted patients were included in this study. The genotype of the HLA-G 14-bp ins/del of patients was determined by polymerase chain reaction (PCR). In particular, the insertion/insertion (ins/ins) genotype was significantly associated with low CMV infection (OR= 0.22, 95% CI: 0.06 0.85; P = 0.02). The deletion/deletion (del/del) and ins/del genotypes were not associated with a lower risk of contracting CMV. Renal transplant recipients with ins/ins genotype have a lower risk of being infected with CMV.
Keywords
CMV; HLA-G polymorphism; Kidney transplantation
Introduction
CMV infection occurs worldwide and is usually asymptomatic in healthy individuals. The CMV is also associated with direct and indirect effects in kidney transplant recipients [1]. Both innate and adaptive immunity are necessary for resistance to CMV infection [2]. Data accumulated in regard to HLA-G expression in chronic viral disease has pointed out a potential role ofHLA-G in virus evasion against immune surveillance [3].
HLA-G molecules act as negative regulators of the immune response and their expression in virus-infected cells may enable them to escape immunosurveillance [4]. HLA-G gene is characterized by low allelic polymorphisms when compared to the classic HLA class-I genes. However, an (ins/del) of 14-bp (rs66554220) in the 3’ untranslated region of the gene was associated with the stability of HLA-G Messenger RNA (mRNA) as well as protein levels [5]. On the other hand, a fraction of HLA-G mRNA transcripts presenting the 14-base insertion can be further processed (alternatively spliced) by the removal of 92 bases from the mature HLA-G mRNA[6], yielding smaller HLA-G transcripts, reported to be more stable than the complete mRNA forms [7].
The importance of HLA-G in the immune system is denoted by its capacity to inhibit immune responses at several levels, acting in different immune cell types. The immunosuppressive HLA-G activity is of interest to some pathogens, since they might use these same mechanisms to escape immune surveillance [8]. In the context of CMV infection in humans, the HLA-G 14-bp ins/del polymorphism may play a role in the natural history of CMV infection and in its complications. A sectional study was performed evaluating the polymorphism of 14- bp of HLA-G gene in renal transplant recipients infected or not by CMVwith the aim of verifying if the genetic polymorphism of this gene is associated with a lower risk of CMV infection in transplanted patients.
Materials and Methods
Study population and clinical samples
To be eligible for the extension study, 106 kidney transplant patients were selected from a university outpatient clinic of nephrology in Brazil. Written informed consent was obtained from all patients and the institutional board approved the protocol. Blood samples were collected and were centrifuged to separate the serum and whole blood. Clinical and laboratory data were obtained by examination of the patient records.
HLA-G genotyping
The HLA-G region containing the polymorphism was amplified by PCR using the forward (5’ - GTGATGGGCTGTTTAAAGTGT- CACC -3’) and reverse (5’ - GGAAGGAATGCAGTTCAGCATGA-3’) primers as described previously [9]. Genotypes were determined by electrophoresis in 4.0% Nusieveagarose gel.
Viral DNA extraction
DNA was extracted from 200μl of serum samples using the QIAamp DNA kit (Qiagen, California, USA) according to the manufacturer’s recommendations. The DNA samples were resuspended in 100μl of buffer AE.
Molecular detection of CMV infection by nested-PCR
For detection of viral DNA in serum sample, a nested-PCR for amplification of a conserved region of CMV of the major immediate early gene was carried out [10]. For the first reaction the primers used were MIE4 (5’- CCAAGCGGCCTCTGATAACCAAGCC-3’) and MIE5 (5’- CAGCACCATCCTCCTCTTCCTCTGG-3’) to amplify one fragment of 435 bp and for the second reaction IE1 (5’-CCAC- CCGTGGTGCCAGCTCC-3’) and IE2 (5’ CCCGCTCCTCCTGAG-CACCC-3’) primers were used to amplify one fragment of 159bp [10]. Positive and negative controls were included in all assays. As a positive control we used the DNA extracted from a stock of CMV AD169 strain and as negative control we used ultra-pure water. The amplified products were detected by direct analysis on 2% agarose gel to visualize the fragment of 159bp.
Statistical analysis
Statistical analysis was conducted using the SPSS version 20.0 software for Windows (SPSS, Inc., Chicago, IL, USA), using a significance level of P = 0.05. For comparison between groups, the Mann-Whitney test and chi-square test were used for continuous nonparametricvariables and categorical variables, respectively. In addition, a polytomous logistic regression model was used to estimate the genetic effect on infection risk (OR≥1.0 was associated with susceptibility and an OR≤1.0 conferred protection). Genotype and allele distributions between groups (CMV+ vs CMV-) were compared by Fisher’s exact test. In order to estimate the effect of the insertion homozygote genotype on CMV infection risk, we performed a logistic regression under the recessive genetic model assumption (Deletion/Deletion=0; Deletion/ Insertion=0; Insertion/Insertion=1). Receptor’s age, sex, ethnicity, as well as ganciclovir and valganciclovir usage was all included as covariates in the regression model.
Results
Characteristics of study subjects
The demographic characteristics and basic laboratory data are listed in Table 1. We found among kidney transplant recipients 35% (37/106) infected with CMV (CMV+) and 65% (69/106) uninfected (CMV-). The average age for CMV+ group was 42.9 and CMV- was 41.2, showing no significant difference (P = 0.59). The CMV+ group was 65% male and CMV -, 57% (P = 0.4). Other factors such as an- cestry, symptomatic profile and type of treatment were analyzed in the infection profile of CMV. No significant differences were observed.
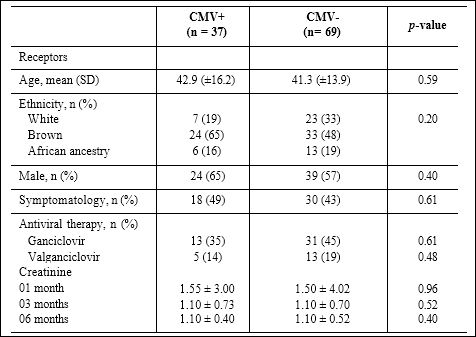
Table 1: Demographic profiles and clinical features.
Comparisons for variables, percentages were compared by Chi-square tests. Comparisons for creatinine were performed by the Mann–Whitney test.
Impact of HLA-G 14-bp polymorphism on CMV infection
Data distribution of genotypes and alleles of the HLA-G 14pb in CMV + kidney transplant patients is shown in Table 2. The total pop- ulation (P = 0.0769) and CMV+ patients (P = 0.3205) were in Har- dy-Weinberg equilibrium, however, CMV- patients deviated from equilibrium (P = 0.0034). A decrease was observed in the frequency of genotype 14-bp ins/ins in positive CMV patients compared to nega- tive CMV patients, so this genotype showed a protective effect (OR = 0.22, 95% CI: 0.06 - 0.85; P = 0.02).
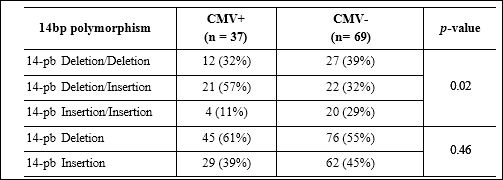
Table 2: Genotypic and allelic distribution for HLA-G 14-bp polymorphism among CMV+ and CMV- renal transplant patients.
Notes: Comparison was performed by Fisher´s exact test.
Discussion
In the context of viral infections, some studies have been conducted reporting HLA-G up regulation associated with HIV [11] HCV [12], HBV [13] HPV [14] and H. pylori [15] infections. Importantly, the reduced immune responses have been associated with soluble HLA-G molecules, which may contribute to the ability of these microorganisms to establish persistent infection in the host. In regard to the overall incidence of CMV infection and disease, there are few studies that address directly this infection in renal transplant patients with HLA-G [16]. Overall, studies have shown that the course of transplant is affected by two HLA-G-mediated mechanisms inhibiting both NK cell and CD8+ T cell-mediated cytolysis as well as suppression of CD4+ T cells all proliferative response [17]. Both mechanisms demonstrate that HLA-G molecules affect main effector cells involved in graft rejection.
The 14-bp insertion / insertion genotype is related to the protection against CMV infection, i.e., patients with 14-bp ins/ins genotype who underwent kidney transplant had a 78% decreased risk of becoming infected by the CMV virus, strengthening the hypothesis that the presence of 14-bp insertion allele favors the reduction of HLA-G expression, thus providing an effective immune response against CMV. HLA-G can modulate the immune response through different mechanisms and favor the viral evasion by subverting host defenses, resulting in the spread of the virus [18]. In many studies, the 14-bp insertion/insertion genotype was related to the low production of the HLA-G molecule [19].
In the same context of kidney allograft recipients infected with CMV [16], the increase of 14-bp deletion/deletion genotype associ- ated with high level of CMV infection was observed. This association was also observed by Zheng et al. [18], suggesting that the 14-bp deletion/deletion genotype and the 14-bp deletion allele can be susceptibility markers to CMV infection in renal allograft recipients. Likewise, it was demonstrated that the 14-bp deletion/deletion polymorphism of HLA-G influences the expression of sHLA-G, in that it is associated with fewer episodes of rejection [20]. Thus, the presence of 14-bp deletion allele provides the graft acceptance by the host since it favors HLA-G expression and inhibits the immune response in the micro environment, consequently supporting the infection and spread of the virus.
Although the HLA-G 14-bp deletion/insertion polymorphism is associated with the expression level of HLA-G [21] modulation of messenger RNA (mRNA) of HLA-G [15], the mechanisms involved in the modulation have not yet been elucidated. The 14-bp deletion allele is associated with the highest magnitude of HLA-G expression because it is associated with the highest stability of the mRNA [19]. On the other hand, the mRNA from the 14-bp insertion allele can undergo an alternative editing process in which 92b are removed, making the mature mRNA less stable, reducing the soluble isoform of HLA-G [18].
In the context of infectious diseases, few studies have been conducted which report on the polymorphism of HLA-G. Considering 14-bp insertion allele specifically, the 14-bp insertion/insertion genotype was associated with increased susceptibility to the virus of human hepatitis B (HBV), since it presents a higher rate of viral replication compared to individuals with the 14-bp deletion/deletion genotype and 14-bp deletion/insertion genotype [13]. Another study related a higher frequency of 14-bp insertion/insertion genotype in African HIV positive patients co-infected with human hepatitis C virus (HCV), suggesting a key role of HLA-G in HIV and HCV co-infection susceptibility [12].
In conclusion, we can assign a less tolerogenic profile to 14-bp insertion/insertion genotype, which directly affects CMV infection in this group of kidney allograft recipients, suggesting the polymorphism of HLA-G 14 bp as a genetic marker of susceptibility to viral infections.
References
- McIntosh M, Hauschild B, Miller V (2016) Human cytomegalovirus and transplantation: drug development and regulatory J Vi- rus Erad 2: 143-148.
- La Rosa C, Diamond DJ (2012) The immune response to human CMV. Future Virol 7: 279-293.
- Donadi EA, Castelli EC, Arnaiz-Villena A, Roger M, Rey D (2011) Implications of the polymorphism of HLA-G on its function, regulation, evolution and disease association. Cell Mol Life Sci 68: 369-
- Xu HH, Shi WW, Lin A, Yan WH (2013) HLA-G 3’ untranslated region polymorphisms influence the susceptibility for human papillomavirus infection. Hum Immunol 74: 1610 5.
- Laaribi AB, Zidi I, Hannachi N, Ben Yahia, Chaouch H, et (2015) Association of an HLA-G 14-bp Insertion/Deletion polymorphism with high HBV replication in chronic hepatitis. J Viral Hepat 22: 835- 841.
- Castelli EC, Mendes-Junior CT, Veiga-Castelli LC, Roger M, Moreau P, et al. (2011) A comprehensive study of polymorphic sites along the HLA-G gene: implication for gene regulation and Mol Biol Evol 28: 3069-3086.
- Jeong S, Park S, Park BW, Park Y, Kwon OJ, et al. (2014) Human leukocyte antigen-G (HLA-G) polymorphism and expression in breast cancer patients. PLoS One 9: e98284.
- Morandi F, Rizzo R, Fainardi E, Rouas-Freiss N, Pistoia V (2016) Recent Advances in Our Understanding of HLA-G Biology: Lessons from a Wide Spectrum of Human Diseases. J Immunol Res 2016:
- Hviid TV, Hylenius S, Hoegh AM, Kruse C, Christiansen OB (2002) HLA-G polymorphisms in couples with recurrent spontaneous abor Tissue Antigens 60: 122-132.
- Tokimatsu I, Tashiro T, Nasu M (1995) Early diagnosis and monitoring of human Cytomegalovirus Pneumonia in patients with adult t-cell leukemia by DNA amplification in serum. Chest 107: 1024-
- Donaghy L, Gros F, Amiot L, Mary C, Maillard A, et al. (2007) Elevated levels of soluble non-classical major histocompatibility class I molecule human leucocyte antigen (HLA)-G in the blood of HIV -infected patients with or without visceral leishmaniasis. Clin Exp Immunol 147: 236-40.
- Silva GK, Vianna P, Veit TD, Crovella S, Catamo E, et (2014) Influence of HLA-G polymorphisms in human immunodeficiency virus infection and hepatitis C virus co-infection in Brazilian and Italian individuals. Infection, Genetics and Evolution 2: 418-423.
- Souto FJ, Crispim JC, Ferreira SC, da Silva AS, Bassi CL, et al. (2011) Liver HLA-G expression is associated with multiple clinical and histopathological forms of chronic hepatitis B virus infection. J Viral Hepat 18: 102-105.
- Yang YC, Chang TY, Chen TC, Lin WS, Chang SC, et (2014) Human leucocyte antigen-G polymorphisms are associated with cervical squamous cell carcinoma risk in Taiwanese women. European Journal of Cancer 50: 469-474.
- Genre J, Reginaldo FP, Andrade JM, Lima FP, da Camara AV, et (2016) HLA-G 14-bp Ins/Ins Genotype in Patients Harbouring Helicobacter pylori Infection: A Potential Risk Factor?. Scand J Immunol 83: 52-57.
- Misra MK, Prakash S, Kapoor R, Pandey SK, Sharma RK, et al. (2013) Association of HLA-G promoter and 14-bp insertion-deletion variants with acute allograft rejection and end-stage renal disease. Tissue Antigens 82: 317-326.
- Rebmann V, Nardi FS, Wagner B, Horn PA (2014) HLA-G as a Tolerogenic Molecule in Transplantation and J Immunol Res 2014: 297073.
- Zheng XQ, Zhu F, Shi WW, Lin A, Yan WH (2009) The HLA-G 14 bp insertion/deletion polymorphism is a putative susceptible factor for active human cytomegalovirus infection in children. Tissue Antigens 74: 317-321.
- Shankarkumar U, Shankarkumar A, Chedda Z, Ghosh K (2011) Role of 14-bp deletion/insertion polymorphism in exon 8 of the HLA-G gene in recurrent spontaneous abortion J Hum Reprod Sci 4: 143-146.
- Torres MI, Luque J, Lorite P, Isla-Tejera B, Palomeque T, et al. (2009) 14-Base pair polymorphism of human leukocyte antigen-G as genetic determinant in heart transplantation and cyclosporine therapy monitoring. Hum Immunol 70: 830-835.
- Haghi M, HosseinpourFeizi MA, Sadeghizadeh M, Lotfi AS (2015) 14-bp Insertion/Deletion Polymorphism of the HLA-G gene in Breast Cancer among Women from North Western Asian Pac J Can- cer Prev 16: 6155-6158.
Citation: Shareef SA, Arnaout RK, Hasnain SM, Al Banyan M, Al-Masri W, et al. (2020) First Report of Rush Immunotherapy for Samsum Ant. J Immuno Immunothe 3: 005.
Copyright: © 2020 Shareef SA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and re- production in any medium, provided the original author and source are credited.


