*Corresponding Author:
Ying Gao,
Department of Comprehensive Internal Medicine, Xinjiang Medical University, Xinjiang, China
Tel: + 86 1399 9815660
E-mail: gaoydct@163.com
Abstract
Background: In this current work we aimed to find the effect of four different single Nucleotide Polymorphisms (SNPs) rs1122608 (SMARCA4), rs2230806 (ABCA1), rs12563308 (ANGPTL3), and rs662799 (APOA5) on Coronary Heart Disease (CHD) in a Han Chinese population in Xinjiang region of China.
Methods: This study involved 914 subjects with 493 CHD patients and 421 healthy controls. The genotype distribution of these SNPs was analyzed and their relations with CHD risk factors were assessed.
Results: No statistical differences were found in genotype and allele distributions of above SNPs between CHD and healthy controls (P>0.05). Serum level of high-density lipoprotein cholesterol (HDL-C) was higher in TT genotype of rs1122608 when compared with GT and GG genotypes (P<0.01) in CHD patients; serum Triglyceride (TG) level was higher in rs662799 GG genotype than GA and AA genotypes (P<0.00); AA of rs662799 was associated with higher HDL-C level compared with other two genotypes (P<0.01); rs12563308 and rs223086 were not associated with any serum lipid traits (P>0.05 for all). Logistic regression analysis showed that the SNPs examined were not related to CHD (p>0.05). Also no association was found between four SNPs with the angiographic severity of CHD patients (p>0.05).
Conclusion: APOA5 rs662799 GG allele is associated with elevated triglyceride and might act as a risk factor for CHD; SMARCA4rs1122608 TT allele and APOA5 rs662799 AA allele are associated with elevated high-density lipoprotein cholesterol levels, and might play a protective role in the development of CHD.
Keywords
Coronary heart disease; Han Chinese; SNPs; Lipids
Introduction
Coronary Heart Disease (CHD), also called coronary artery disease, refers to the cardiovascular disease with a narrowing or blockage of the coronary arteries, which is the major cause of mortality and disability worldwide [1]. Despite significant advances in clinical treatment of CHD, it is still one of the common causes of adult mortality in both developed and developing countries. In China, according to an estimated report by the World Health Organization, more than 700,000 people die from CHD each year [2]. CHD is a complex and multi-factorial disorder involving the interplay of both genetic and environmental factors. Many risk factors such as abnormal plasma lipid concentrations, smoking, diabetes and blood pressure have been proven to be closely associated with the pathogenesis of CHD, which are addressed as “modifiable risk factors” as they can be adjusted by life style changes and therapeutic interventions [3,4]. While lifestyle modification has reduced the mortality rate, the candidate gene approach has provided new insights for discovering diagnostic and therapeutic approaches. The integrative approach of analyzing the disease both clinically and genetically provides the opportunity to disentangle the complex interactions underlying disease pathways [5]. As such, the important role of lipid regulatory genes in CHD pathogenesis has been an essential aspect of research for the past many years [6]. Previous studies have found many lipids related Single Nucleotide Polymorphisms (SNPs) associated with the risk of CHD occurrence. However, the results are inconsistent [2,7-9]. While more and more Met S risk loci have been identified, it has long been noted that genetic variants conferring susceptibility may vary across ethnicities. Among the genes involved in the development of MetS and/or cardiovascular diseases are the apolipoprotein A5 (APOA5), apolipoprotein C1 (APOC1), BRCA1 associated protein (BRAP), BUD13 homolog (BUD13), Cholesteryl Ester Transfer Protein (CETP), lipase A lysosomal acid type (LIPA), Lipoprotein Lipase (LPL), phospholipase C gamma 1 (PLCG1), and ZPR1 Zinc Finger (ZPR1) gene.
An important lipid regulatory gene is BRG1 (also known as SMARCA4) is located about 36 kbs from the LDLR gene.SNP rs1122608 is located in the 30th intron of the SMARCA4 gene, also commonly referred to as BRG1, which has SNPs directly related to dyslipidemia [10]. Zhou et al. [11] showed that SNP rs1122608 (G/T) is related to the increase of LDL-cholesterol. Wang and colleagues [2] also proved that rs1122608 is associated with LDL-cholesterol levels and Triglycerides (TG) levels.
ANGPTL3 gene has been mapped to the 1p31 region, and the SNPs of ANGPTL3 are involved in the metabolic regulation of triglycerides, LDL-C, and HDL-C, as well as atherosclerosis in mice and humans [12]. Li [13] showed that ANGPTL3 rs12563308T haplotype was associated with an increased angiographic severity to coronary artery atherosclerosis.
ABCA1 is a membrane transporter protein involved in creating nascent HDL-C [14]. It plays an essential role incellular free cholesterol and phospholipid secretion from cell membrane to lipid poor apolipoprotein AI [15]. To date approximately 100 gene mutation sites have been reported in ABCA1, among them, rs2230806 (R219K, 107620867C>T) is the most widely studied common missense polymorphism [16]. However, the relationship between rs2230806 and CHD was not consistent in the reported researches.
Apolipoprotein A5 (ApoA5), a protein composed of 366 amino acids and High-Density Lipoprotein (HDL) primarily secreted from the liver is a well-known modulator of circulating TG levels in both fasting and postprandial states [17]. APOA5 polymorphisms have long been reported to be associated with cardiovascular disease and plasma lipid levels. a strong statistical association between HDL-C and LDL-C clinical parameters and APOA5 rs662799 CC and rs3135507 AA genotype was found (p=0.014 and p=0.017, respectively) [18].
In this study, we aim to investigate the association of four different SNPs of lipid metabolism genes, rs1122608(SMARCA4), rs2230806(ABCA1), rs12563308 (ANGPTL3), and rs662799 (APOA5) and their effect on lipids and the severity of CHD in a Chinese Han population in Xinjiang.
Methods
Ethical approval of the study
The present study was approved by the Ethics Committee of First Affiliated Hospital of Xinjiang Medical University (Xinjiang, China). All of the participants provided written informed consent and pro- vided permission for DNA analyses, as well as for the collection of relevant clinical data.
Subjects
This study was carried out in a case–control design. A total of 914Han Chinese subjects (493 of them whom were diagnosed CHD cases and 421 were healthy controls) in Xinjiang region were recruit- ed from the First Affiliated Hospital of Xinjiang Medical University from January 2017 and to December 2019. All subjects were Chinese Han population living in Xinjiang province.CHD was defined as pres- ence of at least one significant coronary arteries, left anterior descend- ing artery, left circumflex or right coronary artery stenosis of >50% luminal diameter based on the coronary angiography .Accordingly, CHD patients were classified as single, double and multivessel ste- nosis patients. The Gensini scores of the control group patients were evaluated according to the Gensini score standard [19].
Exclusion criteria: Patients with incomplete clinical data and pa- tients with one of the following diseases, such as rheumatic heart dis- ease, congenital heart disease, aortic dissection, severe heart failure, cardiogenic shock, malignant arrhythmia, tumor, autoimmune dis- ease, mental disease and patients with liver, kidney or lung dysfunc- tion. Control subjects were selected from volunteers with angiograph- ically normal coronary arteries and had no history of CHD . Coronary angiography in the control individuals was performed for the evalua- tion of chest pain. Some CHD patients had taken lipid lowering med- ications before they were admitted to hospital. A total of 113 (23%) patients used statins and 5 patients used other lipid lowering herbs.
Collection of basic materials
The following basic information was collected: age, gender, total TC, TG, HDL-C andLDL-C, ApoA, ApoB, Lp (a) and glucose levels.
After fasting for 12 hour, a venous blood sample of 5 ml was ob- tained from all participants. 2 ml of the blood sample was collected into glass tubes and used to measure serum lipid levels, another 3 ml was stored in the tubes that contained anticoagulants (4.80 g/Lcitric acid, 14.70 g/L glucose, and 13.20 g/L tri-sodiumcitrate) and used to extract deoxyribonucleic acid (DNA). The levels of serum TC, TG, HDL-C, and LDL-C were determined by enzymatic methods with commercially available kits(RANDOX Laboratories). Serum ApoA1 and ApoB levels were detected by the immunoturbidimetric immuno- assay. The normal values in our Clinical Science Experiment Center were 3.10-5.17 mmol/L for TC, 0.56–1.70 mmol/L for TG, 0.91–1.81 mmol/L, for HDL-C, 2.70–3.20 mmol/L for LDL-C, 1.00–1.78 g/L for ApoA1,0.63–1.14 g/L for ApoB. Diabetes mellitus was diagnosed according to the WHO diagnostic criteria, which are random plasma glucose more than 11.1mmol/L or fasting plasma glucose 7.0 mmol/L or higher [20]. Hypertension was defined as a systolic blood pres- sure of 140 mmHg or greater, and/or a diastolicblood pressure of 90 mmHg or higher [20].
Genotyping
Genomic DNA was extracted from leucocytes of venous blood using the phenol-chloroform method. Genotyping of the SNPs was accomplished by the Snapshot technology platform in the Center for Human Genetics Research, Shanghai Genesky Bio-Tech Co. Ltd.
The restriction enzymes for the loci were SAP (Promega) and Exonucleasel (Epicentre). The sense and antisense primers were 5’-CAACACTGCGAGGCGGGTAC-3′and 5’-CAACACTGC- GAGGCGGGTAA-3 for the rs1122608SNP and 5’-TAGTGAAG- CAATCTAATTATGTTTTACGAATTGTGT-3′ and 5’-TAGT- GAAGCAATCTAATTATGTTTTACGAATTGTGC-3′ for the rs12563308 SNP; 5’-CTCTGCTGCAGCCAGTTTCTACC-3′and 5’-CTCTGCTGCAGCCAGTTTCTGCT-3 for the rs2230806 SNP, 5’-CCCAGGAACTGGAGCGAAACTG-3′and 5’-CCCAGGAACTGGAGCGAAACTA-3 for the rs662799 SNP respectively. In or- der to identify the most likely relative lipid SNP candidate genes, we accessed to http://www.ncbi.nlm.nih.gov/SNP database and selected rs1122608(SMARCA4), rs2230806(ABCA1), rs12563308 (ANGPTL3), and rs662799 (APOA5) genes asour candidate genes association with blood lipids and CHD.
Statistical analysis
The data analysis was performed using SPSS version 25.0 for Windows. Each SNP was coded as 0, 1, or 2 depending on the num- ber of CHD risk alleles in the patient. The genotype distribution was assessed by Chi-sequare test. The differences in genotype and allele frequencies between different groups were also examined by Hardy- Weinberg Equilibrium (HWE) test [21]. Student’s t test or the analy- sis of variance was used to compare the clinical parameters between cases and controls. Qualitative variables were reported as frequencies and percentages and evaluated by Chi-square test. Multivariate lo- gistic regression analyses were performed for SNPs and other risk factors associated with CHD and the Odds Ratio (OR) and 95% Con- fidence Interval(CI) were calculated to evaluate the contribution of the major risk factors. Through counting DNA sequencing data, the genotype and allele frequencies can be estimated. The distinction be- tween studied groups were analyzed by Pearson’s x2 test. The thresh- old for significance was set at P<0.05.
Results
Tables 1-7 are shown in below.
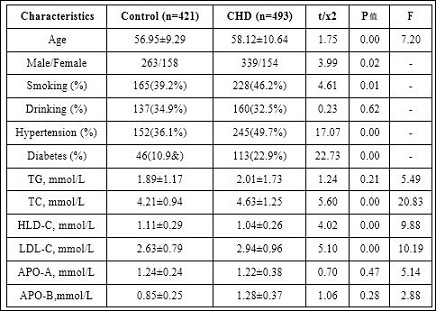
Table 1: General characteristics of participants.
Note: Continuous variables are expressed as mean +− SD. Categorical variables are expressed as percentages. The P value of the continuous variables was calculated by the independent samples t test. The P value of the categorical variables was calculated by χ2 test. Abbreviations: HDL-C: high-density lipoprotein-cholesterol; LDL-C: low-density lipoprotein-cholesterol; TC:total cholesterol; TG: triglyceride. APO-A: Apolipoprotein A. APO-B: Apolipoprotein B.
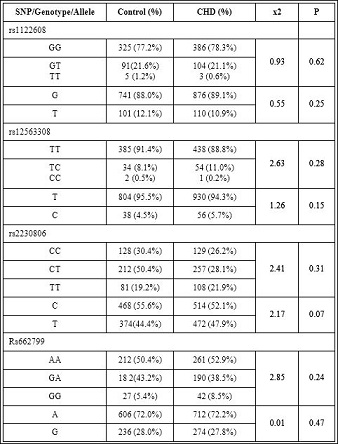
Table 2: Genotypic and allelic distributions of SNPs between control and CHD group (P>0.05).
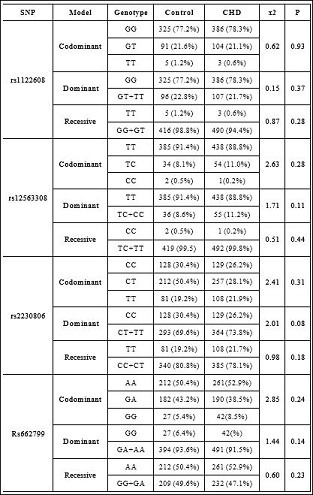
Table 3: Codominant, Dominant, Recessive models of SNPs between control group and CHD group.
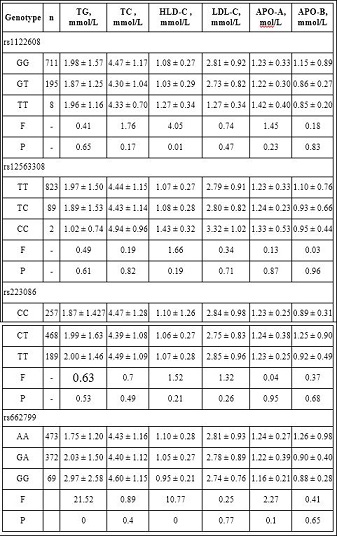
Table 4: Association of SNPs and serum lipid levels in CHD group.

Table 5: Multivariable logistic regression analyses of the major confounding factors for CHD.
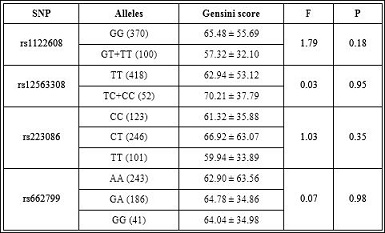
Table 6: Gensini score of SNPs.
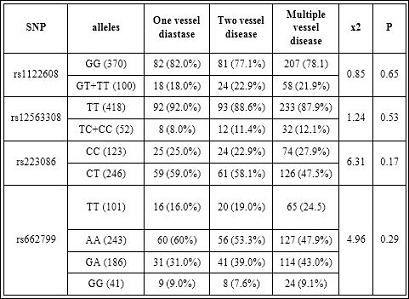
Table 7: Effect of SNPs on angiographic severity of CHD.
Discussion
In this study we investigated the associations between SNPs of lipid metabolizing genes and their relations to 493 diagnosed CHD cases and 412 healthy controls from Han Chinese population in Xinjiang region of China. We chose four different SNPs related with lipid metabolism, they are rs1122608 in SMARCA4, rs2230806 in ABCA1, rs12563308 in ANGPTL3, and rs662799 APOA5 and evaluated whether they were associated with lipid levels and the risk of CHD in Han Chinese population. Our results showed no significant differences of these SNPs and their genotypic and allelic distributions between control and CHD group (P>0.05) (Table 2). In other words there is no statistically significant association between above four SNPs and CHD risk. Serum level of HDL-C was higher in TT genotype of rs1122608 when compared with GT and GG genotype (P<0.01); serum Triglyceride (TG) levels was higher in rs662799 GG genotype than GA and AA genotype (P<0.00); AA of rs662799 had higher HDL-C level compared with the other two genotypes (P<0.01) ; rs12563308 and rs223086 were not associated with any serum lipid traits (P>0.05 for all) (Table 4). Following multivariate adjustments for the confounders, such as age, sex, smoking, hypertension and diabetes, rs1122608,rs2230806, rs12563308,and rs662799 were still not related with CHD (P>0.05 ). Also no association was found between four SNPs with gensini score and the angiographic severity of CHD patients (P>0.05 ) (Table 5).
Genome-wide association studies have identified rs1122608 SNP, located in intron 30 of BRG1/SMARCA4, as a risk variant for CHD [22]. Moreover, much evidence has verified that rs1122608 is related to CHD independently of lipid profiles [23,24]. A report of this SNP on Iranian populations showed that intron 30 of SMARCA4 gene include rs1122608 was associated with a strong protective effect against CHD [25]. Fujimaki et al. [26] performed a case-control study and that rs1122608 of SMARCA4 (P<0.0305; dominant model; odds ratio, 0.86) is a risk factor of hypertension in Japanese individuals. Guo X, Wang X, et al. [27] found significant differences in glucose concentrations of rs1122608 different genotype, but no significant association was found between rs1122608 polymorphism and Coronary heart disease or lipid metabolism in Han population living in Xi’an city. In a different Chinese Han study population, it has been pointed out by Chen QF, et al. [28] that the mutant GT and TT genotypes and minor T allele of rs1122608 are positively correlated with the risk of AMI..
Another study showed, rs1122608 was associated with a higher risk of revascularization of cardiovascular complications in patients with CAD confirmed by coronary angiography [29]. Meta-analyses performed for rs11206510 and rs1122608 showed that two SNPs were associated with CHD in Caucasians but not in Asians [30]. Ma H [31] also found rs1122608 in SMARCA4 seemed to have strong protective effects on the hypertension. Above results showed the controversy effect of this SNF on lipids and CHD.
Our study showed TT genotype of rs1122608 was associated with higher HDL-C levels, therefore may act as a protective factor, however this theory should be proven further due to biographical and ethical backgrounds of our samples (Table 4).
Angiopoietin like 3 gene (ANGPTL3) encodes a member of a family of secret proteins that function in angiogenesis which is involved in the metabolic regulation of triglycerides, LDL-C, and HDL-C, as well as atherosclerosis in mice and humans [32]. Previous GWASes also reported that some ANGPTL3 polymorphisms were associated with serum TG and TC levels [33]. rs12563308 SNP belongs to ANGPTL3 gene family, but little is known about the association of the ANGPTL3 rs12563308 Single Nucleotide Polymorphisms (SNPs) with serum lipid levels and the risk of CHD. Li [34] showed that ANGPTL3 rs12563308 SNPs were not associated with all of seven serum lipid traits in the controls,but rs12563308T haplotype was associated with an increased angiographic severity to coronary artery atherosclerosis. Gong Q at el. [35] explored associations between genetic variants of the ANGPTL3 gene and susceptibility to ischemic stroke in a large-scale case–control study in a Chinese population. They found that rs12563308 were significantly associated with susceptibility to ischemic stroke. They also pointed out that carriers of the minor allele of SNP rs12563308 had significantly lower levels of TC and LDL-C. A study in a Finish population showed that subjects who carried ANGPTL3 sequence variants rs12563308 (n=4) and rs199772471 (n=1) had abnormally high TC and LDL-C concentrations [36].
In our study, rs12563308 was not associated with any serum lipid traits (P>0.05 for all).Also no association was found between rs12563308 with gensini score and the angiographic severity of CHD patients (P>0.05 ) (Tables 6 and 7). However, the reason for these discrepancies in different investigations is still unclear.
The SNP rs2230806 is located in ATP-binding cassette transporter A1 (ABCA1) gene. ABCA1 is a membrane transporter protein that plays an essential role in the effelux of cholesterol from peripheral tissues back to the liver, thus has a crucial role in removing intracellular cholesterol and plays a protective role against atherosclerosis [37]. In recent years, a number of studies have shown that the distribution frequency of specific ABCA1 gene SNPs in different regions and populations is significantly different, therefore the effects on plasma lipid levels and the incidence and severity of CHD are also different [38]. Even the same ABCA1 gene SNPs have similar or opposite effects [39,40]. Therefore, genetic polymorphisms in this gene may alter the susceptibility or the improvement of CHD. Fan Q at el. [41] showed in a meta Analysis of a total of 34,348 subjects (14,085 CAD cases and 20,263 healthy controls) that there is a significant association between rs2230806 polymorphism and the risk of CAD under different genetic models: an allelic genetic model (OR=0.745, 95% CI = 0.687–0.809, P < .001), a recessive genetic model (OR = 0.683, 95% CI = 0.603–0.774, P < .001), a dominant genetic model (OR = 0.703, 95% CI = 0.633–0.781, P < .001), a homozygote genetic model (OR = 0.573, 95% CI = 0.488–0.672, P < .001), and a heterozygote genetic model (OR = 0.761, 95% CI = 0.693– 0.837, P < .001), In addition, statistical result showed rs2230806 is significantly associated with CAD in Asian population, marginally significant in Caucasian and not significant in other group. The reason for racial difference phenomenon may be attributed to allele frequency and other factors, such as lifestyle discrepancy and so on. Ma et al.[42] indicated rs2230806 is a protective factor for CAD risk both in Asians (OR = 0.76, 95% CI = 0.68–0.85) and Caucasians (OR = 0.89, 95% CI = 0.81–0.99. Wang F at el. [43] study results showed that AA genotype of the rs2230806 polymorphism had higher levels of TC, LDL-C and uric acid than those with GA genotype (p<0.05 for all) but no associations betweenrs2230806 polymorphism and severity of CAD was detected.
Our results also found no association between rs2230806 genotype and severity of CHD or with serum lipids levels (Tables 4,6,7).
Apo-lipoprotein A5 (ApoA5) is part of VLDL, HDL and CM and is a major regulator of blood TG and HDL-C through interaction with LDLR [44]. Rs662799 was a polymorphism located in APOA5 gene and is the most studied SNP in the promoter region of this gene [45]. The allele frequency of rs662799 in HapMap database was 1.7, 13.3.26.7 and 28.9% in European, African, Chinese and Japanese respectively. Chen H et al. [46] found a significant association of the SNP rs662799 in APOA5 genes with CAD. Valente-Frossard TNS [47] showed in their study that genotypes of the APOA5 rs662799 were not associated with lipid levels. Another study of Korean metabolic syndrome subjects showed that SNP rs662799 in the APOA5 gene was associated with increased risk of metabolic syndrome and its components, especially elevated TG and low levels of HDL-C [48]. Hsu LC [49] also found out TC + CC genotype of rs662799 is associated with high serum triglyceride and low LDL and BMI levels in the Han Chinese population in Taiwan.
In our study we also found ApoA5 rs662799 was associated with elevated TG in Han Chinese subjects in Xinjiang. The same results was reported in a study conducted in India where the rs662799 was associated with 19% increase in serum TG levels [50], similar TG raising effect of this SNP has also been reported in Chinese people other than Xinjiang region and Paksitani subjects [51-53]. Therefore, ApoA5 rs662799 might play in TG metabolism in Han Chinese people in Xinjiang.
To sum up, in our study, we confirmed polymorphisms of rs1122608(SMARCA4), rs2230806(ABCA1), rs12563308 (ANGPTL3) and rs662799( APOA5) are not different between CHD patients and normal control subjects in Xinjiang Han Chinese population, suggesting that these SNPs and their genotypes and alleles may not be associated with CHD in HanChinese population in Xinjiang. Among these SNPs rs1122608 (SMARCA4) TT allele and rs662799 (APOA5) AA allele were associated with elevated HDL-C levels meanwhile rs662799 (APOA5) GG alelle was also associated with elevated TG levels. According to our results we also concluded these SNPs have no affect on the severity of CHD. GG genotype in rs662799 might be related to the elevated TG levels in CHD patients, therefore may act as a risk factor for CHD, TT genotype of rs1122608 and AA genotype of rs662799 are related with higher HDL-C levels, therefore may play a protective role in the development of CHD in HanChinese population in Xinjiang region.
Investigating these SNPs should use more clinical data with bigger samples. Our current research is fundamental; further functional studies and larger population‐based prospective studies are required to understand the genetic factors underlying CHD.
Declaration
Ethics approval and consent to participate
The present study was approved by the EthicsCommittee of First Affiliated Hospital of Xinjiang Medical University (Xinjiang, China). All of the participants provided written informed consent and provided permission for DNA analyses, as well as for the collection of relevant clinical data.
Funding
This study is supported by National Natural Science Foundation of China in 2017 (No. 81660040).
Authors’ Contributions
Ying Gao contributed to the study design, Shajidan Abudureyimu and Palida Abulaiti performed the data analysis, Shajidan Abudureyimu , Zhi Xing and Hui Li prepared the manuscript, Sha Sha Liu and Wen Li collected the data. All authors read and approved the final manuscript.
Acknowledgments
We are grateful to the patients and control subjects for their participation in this study. We also thank the clinicians and hospital staff who obtained the blood samples and performed data collection for this study.
References
- Chen Q, Li L, Yi J, Huang K, Shen R, et al. (2020) Waist circumference increases risk of coronary heart disease: Evidence from a Mendelian randomization Mol Genet Genomic Med 8: e1186.
- Wang Y, Wang L, Liu X, Zhang Y, Yu L, et al. (2014) Genetic Variants Associated with Myocardial Infarction and the Risk Factors in Chinese PLoS One 9: e86332.
- Kessler T, Vilne B, Schunkert H (2016) The impact of genome‐wide association studies onthe pathophysiology and therapy of cardiovascular dis Embo Molecular Medicine 8: 688-701.
- Spiller W, Jung KJ, Lee JY, Jee SH (2020) Precision Medicine and Cardiovascular Health: Insights from Mendelian Randomization Analyses. Korean Circ J 50: 91-111.
- Jian Y, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, et (2011) Genome partitioning of genetic variation for complex traits using common SNPs. Nature Genetics 43: 519-525.
- Voight BF, Kang HM, Ding, J Palmer CD, Sidore C, et al. (2012) The Metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric PLoS Genet 8: e1002793.
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, et (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466: 707-713.
- Kathiresan S, Manning AK, Demissie S, D’Agostino RB, Surti A, et al. (2007) A genome-wide association study for blood lipid phenotypes in the Framingham Heart BMC Med Genet 8: S17.
- Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, et al. (2013) Large-scale association analysis identifies new risk loci for coronary artery Nat Genet 45: 25-33.
- Ma H, He Y, Bai M, Zhu L, He X, et (2019) The genetic polymorphisms of ZC3HC1 and SMARCA4 are associated with hypertension risk. Mol Genet Genomic Med 7: e942.
- Zhu H, Tucker HM, Grear KE, Simpson JF, Manning AK, et al. (2007) A common polymorphism decreases low-density lipoprotein receptor exon 12 splicing efficiency and associates with increased Hum Mol Genet 16: 1765–1772.
- Gong Q, Ye L, Gui H, Liu J, Li H, et (2019) Association study of genetic variants of the ANGPTL3 gene and susceptibility to ischemic stroke. Neuropsychiatr Dis Treat 15: 3015-3020.
- Li WJ, Yin RX, Cao XL, Chen WX, Huang F, et al. (2018) DOCK7- ANGPTL3 SNPs and their haplotypes with serum lipid levels and the risk of coronary artery disease and ischemic Lipids Health Dis 17: 30.
- Oldoni F, van Capelleveen JC, Dalila N, Wolters JC, Heeren J, et (2018) Naturally occurring variants in LRP1 (low‐density lipoprotein receptor‐related protein 1) affect HDL (high‐density lipoprotein) metabolism through ABCA1 (ATP‐binding cassette A1) and SR‐B1 (scavenger receptor class B Type 1) in humans. Arterioscler Thromb Vasc Biol 38: 1440-1453.
- Yan H, Cheng L, Jia R, Yao H, Wu H, et al. (2019) ATP-binding cassette sub-family a member1 gene mutation improves lipid metabolic abnormalities in diabetes mellitus. Lipids Health Dis 18: 103.
- Du W, Hu Z, Wang L, Li M. Zhao D, et al. (2020) ABCA1 Variants rs1800977 (C69T) and rs9282541 (R230C) Are Associated with Susceptibility to Type 2 Public Health Genomics 23: 20-25.
- Fahrioğlu U, Ergören MÇ (2018) The Association Between APOA5 Gene Polymorphisms and Plasma Lipids in the Turkish Cypriot Population: A Possible Biomarker for Preventing Cardiovascular Biochem Genet 56: 176-187.
- Lin E, Kuo PH, Liu YL, Yang AC, Tsai SJ (2017) Detection of susceptibility loci on APOA5 and COLEC12 associated with metabolic syndrome using a genome-wide association study in a Taiwanese popula- Oncotarget 8: 93349-93359.
- Nurkalem Z, Hasdemir H, Ergelen M, Aksu H, Sahin I, et al. (2010) The relationship between glucose tolerance and severity of coronary artery disease using the Gensini Angiology 61: 751-755.
- Alberti K, Zimmet P (1999) Definition, diagnosis and classification of diabetes mellitus and its complications: report of a WHO Diabet Med 15: 539-553.
- Chalmers J, MacMahon S, Mancia G, Whitworth J, Beilin L, et al. (1999) 1999 World Health Organization-International Society of Hypertension Guidelines for the management of hypertension. Guidelines sub-committee of the World Health Clin Exp Hypertens 21: 1009-1060.
- Shi YY, He L (2005) SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 15: 97-98.
- Martinelli N, Girelli D, Lunghi B, Pinotti M, Marchetti G, et al. (2010) Polymorphisms at LDLR locus may be associated with coronary artery disease through modulation of coagulation factor VIII activity and independently from lipid Blood 116: 5688-5697.
- van de Woestijne AP, van der Graaf Y, de Bakker PI, Asselbergs FW, de Borst GJ, et (2014) LDL-c-linked SNPs are associated with LDL-c and myocardial infarction despite lipid-lowering therapy in patients with established vascular disease. Eur J Clin Invest 44: 184-191.
- Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, et al. (2009) Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet 41: 56-65.
- Jamaldini SH, Babanejad M, Mozaffari R, Nikzat N, Jalalvand K, et al. (2014) Association of polymorphisms at LDLR locus with coronary artery disease independently from lipid Acta Med Iran 52: 352-359.
- Fujimaki T, Oguri M, Horibe H, Kato K, Matsuoka R, et al. (2015) Association of a transcription factor 21 gene polymorphism with hypertension. Biomed Rep 3: 118-122.
- Guo X, Wang X, Wang Y, Zhang C, Quan X, et al. (2017) Variants in the SMARCA4 gene was associated with coronary heart disease susceptibility in Chinese han population. Oncotarget 8: 7350-7356.
- Chen QF, Wang W, Huang Z, Huang DL, Li T, et (2018) Correlation of rs1122608 SNP with acute myocardial infarction susceptibility and clinical characteristics in a Chinese Han population: A case-control study. Anatol J Cardiol 9: 249-258.
- Wirtwein M, Melander O, Sjőgren M, Hoffmann M, Narkiewicz K, et al. (2017) Relationship between selected DNA polymorphisms and coronary artery disease complications. Int J Cardiol 228: 814-820.
- Zhang L, Yuan F, Liu P, Fei L, Huang Y, et al. (2013) Association between PCSK9 and LDLR gene polymorphisms with coronary heart disease: Case-control study and meta-analysis. Clin Biochem 46: 727-732.
- Ma H, He Y, Bai M, Zhu L, He X, et (2019) The genetic polymorphisms of ZC3HC1 and SMARCA4 are associated with hypertension risk. Mol Genet Genomic Med 7: 942.
- Tikka A, Jauhiainen M (2016) The role of ANGPTL3 in controlling lipoprotein metabolism. Endocrine 52: 187-193.
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, et (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466: 707-713.
- Li WJ, Yin RX, Cao XL, Chen WX, Huang F, et al. (2018) DOCK7ANGPTL3 SNPs and their haplotypes with serum lipid levels and the risk of coronary artery disease and ischemic Lipids Health Dis 17: 30.
- Gong Q, Ye L, Gui H, Liu J, Li H, et al. (2019) Association study of genetic variants of the ANGPTL3 gene and susceptibility to ischemic stroke. Neuropsychiatr Dis Treat 15: 3015-3020.
- Tikka A, Metso J, Jauhiainen M (2017) ANGPTL3 serum concentration and rare genetic variants in Finnish population. Scand J Clin Lab Invest 77: 601-609.
- Sundar PD, Feingold E, Minster RL, DeKosky ST, Kamboh MI (2007) Gender-specific association of ATP-binding cassette transporter 1 (ABCA1) polymorphisms with the risk of late-onset Alzheimer’s disease. Neurobiol Aging 28: 856-862.
- Smirnov GP, Malyshev PP, Rozhkova TA, Zubareva MY, Shuvalova YA, et al. (2018) [The effect of ABCA1 rs2230806 common gene variant on plasma lipid levels in patients with dyslipidemia]. Klin Lab Diagn 63: 410-
- Karimian M, Momeni A, Farmohammadi A, Behjati M, Jafari M, et al. (2020) Common gene polymorphism in ATP-binding cassette transporter A1 and coronary artery disease: A genetic association study and a structural analysis. J Cell Biochem 121: 3345-3357.
- Mokuno J, Hishida A, Morita E, Sasakabe T, Hattori Y, et al. (2015) ATP-binding cassette transporter A1 (ABCA1) R219K (G1051A, rs2230806) polymorphism and serum high-density lipoprotein cholesterol levels in a large Japanese population: cross-sectional data from the Daiko Endocr J 62: 543.
- Fan Q, Zhu Y, Zhao F (2020) Association of rs2230806 in ABCA1 with coronary artery disease: An updated meta-analysis based on 43 research Medicine (Baltimore) 99: 18662.
- Ma X-Y, Liu J-P, Song Z-Y (2011) Associations of the ATP-binding cassette transporter A1 R219K polymorphism with HDL-C level and coronary artery disease risk: A meta-analysis. Atherosclerosis 215: 428-434.
- Wang F, Ji Y, Chen X, Song Y, Huang S, et al. (2019) ABCA1 variants rs2230806 (R219K), rs4149313 (M8831I), and rs9282541 (R230C) are associated with susceptibility to coronary heart disease. J Clin Lab Anal 33: 22896.
- Li R-K, Guo J (2014) Single nucleotide variances can account for loss of microRNA function: The emerging cross talk between genetics and epi J Am Coll Cardiol 64: 278-280.
- Pennacchio LA, Olivier M, Hubacek JA, Cohen JC, Cox DR, et (2001) An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science 294: 169-173.
- Chen H, Ding S, Zhou M, Wu X, Liu X, et al. (2018) Association of rs662799 in APOA5 with CAD in Chinese Han population. BMC Cardiovasc Disord 18: 2.
- Valente-Frossard TNS, Cruz NRC, Ferreira FO, Belisario AR, Pereira BM, et al. (2020) Polymorphisms in genes that affect the variation of lipid levels in a Brazilian pediatric population with sickle cell disease: rs662799 APOA5 and rs964184 ZPR1. Blood Cells Mol Dis 80: 102376.
- Oh SW, Lee JE, Shin E, Kwon H, Choe EK, et (2020) Genome-wide association study of metabolic syndrome in Korean populations. PLoS One 15: 0227357.
- Hsu L-C, Hsu L-S, Lee T-H (2019) Association of apolipoprotein A1 and A5 polymorphisms with stroke subtypes in Han Chinese people in Gene 684: 76-81.
- Chandak RG, Ward KJ, Yajnik CS, Pandit AN, Bavdekar A, et al. (2006) Triglyceride associated polymorphisms of the APOA5 gene have very different allele frequencies in Pune, India compared to BMC Med Genet 7: 1-6.
- Zhu WF, Wang CL, Liang L, Shen Z, Fu JF, et (2014) Triglyceride-raising APOA5 genetic variants are associated with obesity and non-HDL-C in Chinese children and adolescents. Lipids Health Dis 13: 1-7.
- Shahid SU, Shabana NA, Cooper JA, Rehman A, Humphries SE (2017) Common variants in the genes of triglyceride and HDL-C metabolism lack association with coronary artery disease in the Pakistani subjects. Lipids Health Dis 16: 24.
Citation: Abudureyimu S, Abulaiti P, Xing Z, Li H, Liu SS, et al. (2021) The Effect of Four Different Single Nucleotide Polymorphism Son Coronary Heart Disease in a Han Chinese Population in Xinjiang Region. J Cardio Cardiovasc Med 5: 025.
Copyright: © 2021 Abudureyimu S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.


